Users can access the SNObase by the page in http://www.nitrosation.org . Instances about S-nitros(yl)ation could be request by searching the database with different types, including Uniprot ID , S-nitrosation protein (Gene Symbol), S-nitrosation protein (description), S-nitrosation protein (alias), literature title , species, model, related pathophysiological process , etc.
There are some examples as follows:
1, users can know if a protein is an S-nitros(yl)ation target by searching with its Uniprot ID, official gene symbol, protein description or alias. The most exactly results will be get when using Uniprot ID, for it is species specifically.
2, users can know which proteins can be S-nitros(yl)ated in the experimental model or pathophysiological processes they are interested in by searching with the corresponding type.
3, users can request S-nitros(yl)ation targets in a paper searching with literature title. It is convenient to get all S-nitros(yl)ation targets in S- nitrosoproteome works especially.
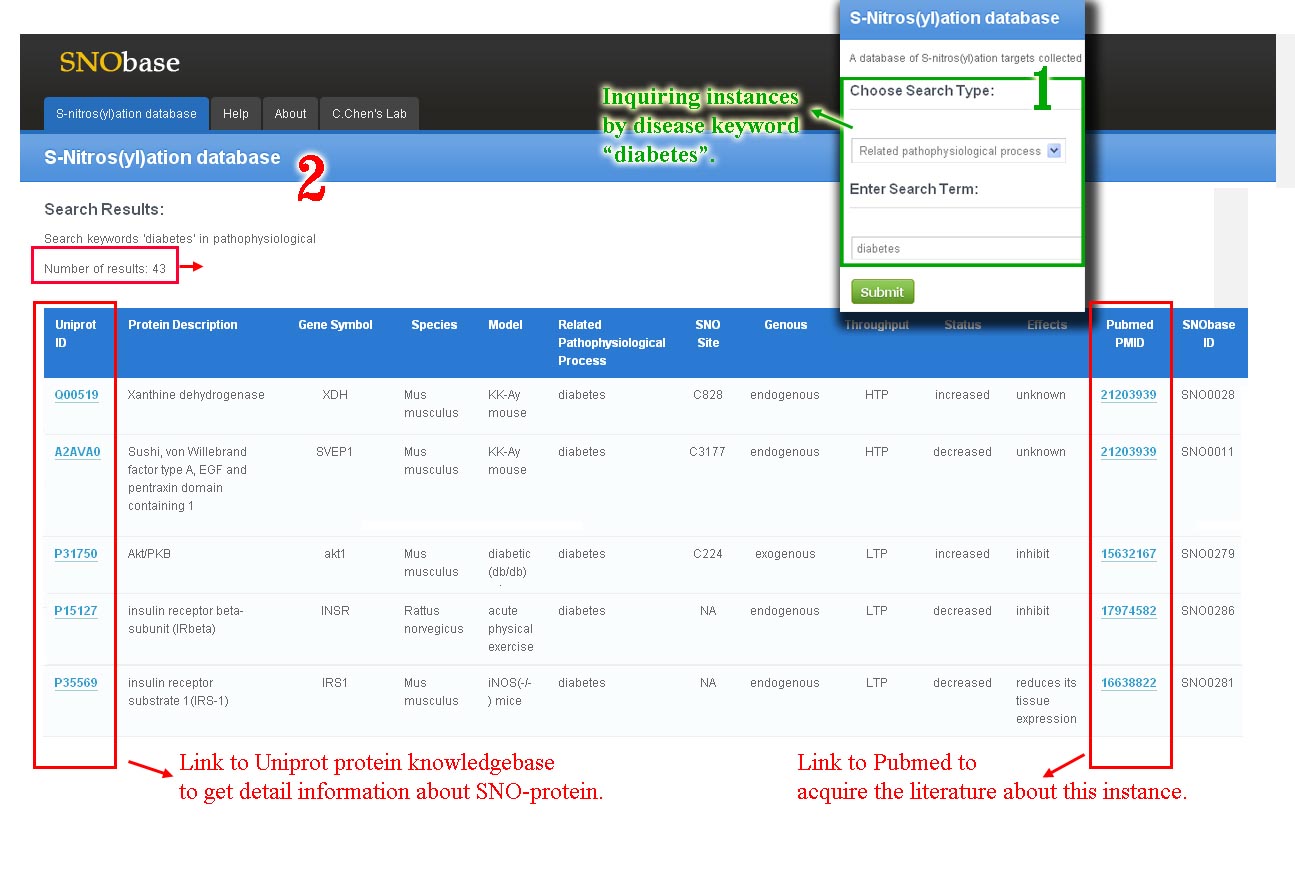
The page retrived is shown as follow. In this example, we search the keywords “diabetes” in the SNObase with the type “ related pathophysiological process ”. 43 instances were shown in the results table. In each insatnce, the Uniprot IDs and Pubmed IDs are hyperlinked to the Uniprot protein knowledgebase and Pubmed database respectively.